Part 1 – Cycles and distribution.
The following is an excerpt from ‘African swine fever in wild boar populations – ecology and biosecurity’ created by the Food and Agriculture Organisation of the United Nations, World Organisation for Animal Health and the European Commission.
African swine fever is a disease of pigs, which was originally associated with the ecological niche of the ticks of the genus ornithodoros and the common warthog – phacochoerus africanus – in sub-Saharan Africa.
Warthogs and ticks, which naturally co-inhabit burrows, can sustain the transmission cycle of this virus for an unlimited time.
It is a well-established natural host–vector–pathogen system, the sylvatic transmission cycle of ASF, with a distribution restricted to parts of the African continent.
Warthogs are naturally resistant to the effects of the African swine fever virus and do not usually develop clinical disease.
Animals are infected when piglets and develop life-long immunity.
In Africa, the virus has shown a trend to shift towards a more anthropogenic cycle – see Figure 1, cycle 2 – in which domestic pigs instead of warthogs assumed the role of an epidemiological reservoir, with the occasional involvement of ornithodoros ticks.
In the past, this kind of transmission cycle was also reported from the Iberian Peninsula.
Again, in Africa, driven by the growing human population and increasing numbers of domestic pigs, ASF spread to areas where it had never occurred naturally before.
In these new areas, its transmission cycle no longer involves ticks or warthogs – see Figure 1, cycle 3.
The virus spread in domestic pigs is facilitated by human activity.
Movements of animals due to trade, the sale of infected meat and free-range pig farming are the main risk factors in this system.
A similar, purely domestic, pig cycle, has also evolved in the Caucasus since 2007, when the genotype II virus was first introduced in Georgia.
Thereafter, it has spread northwards, primarily in the domestic pig population, moving from the Caucasian countries to the Russian Federation, Belarus and Ukraine, and then to other European countries.
Finally, the most recent step in the evolution of the biological cycle of ASFV and its geographical spread is related to the formation of the ‘wild boar-habitat cycle’ – see Figure 1, cycle 4 – which has developed in northern and eastern Europe.
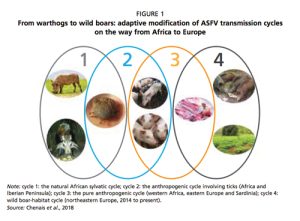
Figure 1: From warthogs to wild boar, adaptive modification of ASFV transmission cycles on the way from Africa to Europe.
This novel host–pathogen–environment system has steadily expanded the range of ASF in Europe, facilitated by the exceptional stability and resilience of ASFV in the environment and infected carcasses of animals.
This cycle is characterised by the continuous presence of the virus in the affected wild boar populations, which represents a serious challenge for the pig production sector and wildlife management authorities, as well as hunters.
In the past four years, ASF has become endemic in wild boar over remarkably large areas and the scale of the problem poses a major threat to the European pig production sector.
Characteristics of the ASF virus circulating in Europe and Asia
ASF is caused by a DNA virus belonging to the asfarviridae family. It affects only those species belonging to the suidae family.
In Europe, the sole susceptible species are domestic pigs and wild boar.
They show similar clinical signs and have similar case fatality rates.
Though a total of 23 genotypes of the virus are known to circulate in Africa, only two of them currently occur in Europe.
Genotype II spread extensively in eastern Europe from 2007, while genotype I has been reported in Sardinia Italy only.
Most recently, genotype II ASFV was introduced and spread over most of China, and from 2018 to 2019 its occurrence range expanded to Mongolia, Vietnam, Cambodia and likely, other countries of the region.
The Genotype II virus now circulating in Europe and Asia has a very high case fatality rate in almost any infected pig, irrespective of whether they are wild or domestic.
The genetic structure of ASFV is rather stable and thus the use of molecular epidemiology for tracing back the origin of the virus is of limited use.







